Oberlin Alumni Magazine
My Cousin the Comb Jelly
November 15, 2023
Annie Zaleski

Animal evolution is one of those major topics you probably learned about in grade-school science class. And it’s a big one to consider: How is it that humans are related to all other living beings on Earth? Not just our obvious relatives like primates, but also the fantastical creatures you might find in a deep ocean dive?
Darrin Schultz ’13, the lead author of a paper published in Nature in May 2023, is taking our understanding of animal evolution even deeper. Using cutting-edge DNA sequencing methods, Schultz and his team analyzed various animal genomes—the unique set of genes that serve as the building blocks of life. Their research concluded that comb jellies—tiny, oval-shaped marine invertebrates known for their iridescent rainbow sheen—are the likely sibling group (in genetic terms, the oldest relation) of all animals. Their findings offer implications for our understanding of how animals are related to one another—and when they started to become more complex.
“There are a lot of new things that we’re really excited to share with other evolutionary biologists,” Schultz says via Zoom from Austria, where he’s a postdoctoral researcher in the University of Vienna’s department of neuroscience and developmental biology. “This paper generates a lot of new hypotheses.”
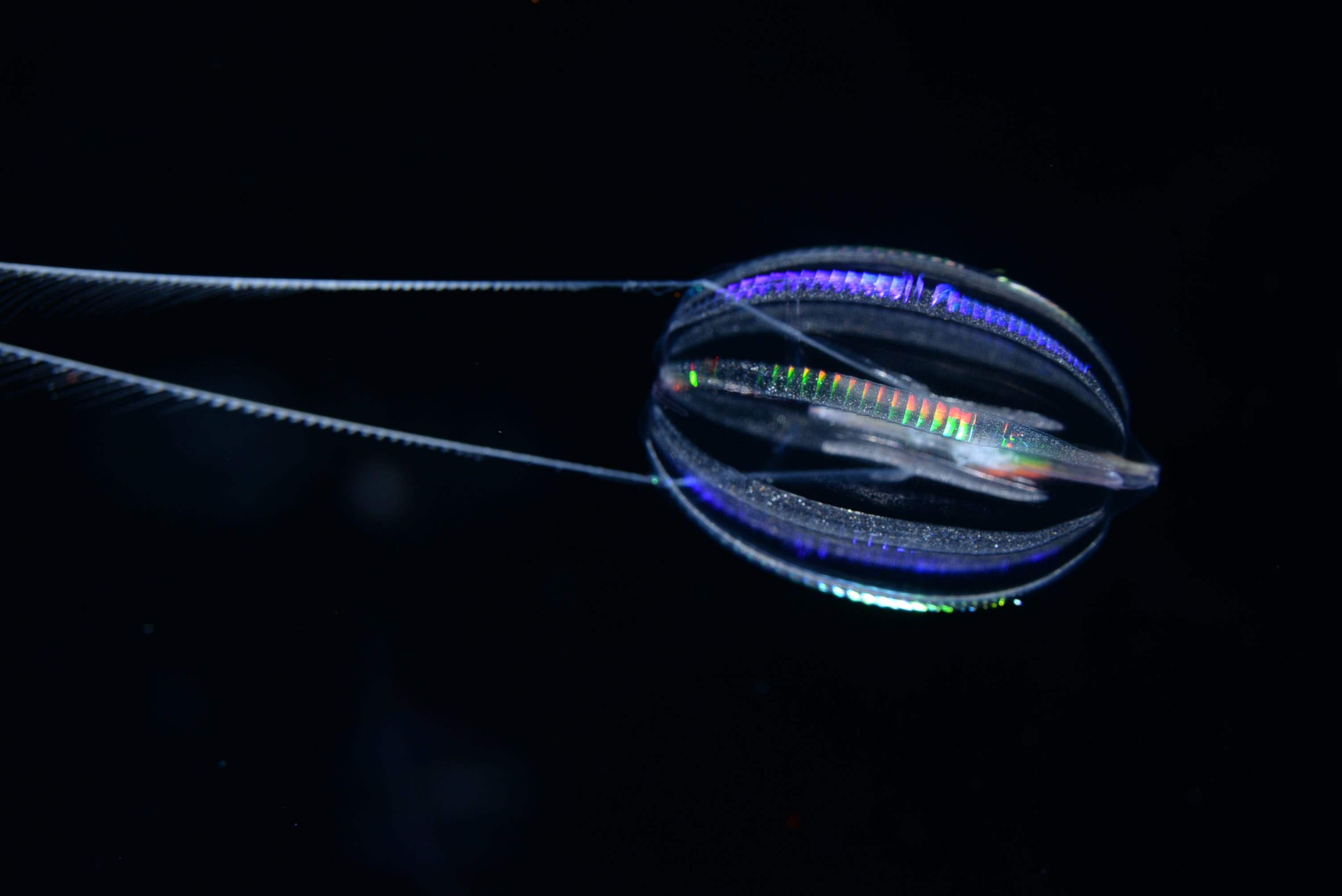
For decades, scientists thought that sponges—superficially simple organisms that lack neurons and muscles— were the sibling group of all animals. This meant sponges had split off onto their own unique evolutionary path before other animal species. However, a groundbreaking 2008 study sparked debate within the scientific community by proposing that ctenophores—also called comb jellies—were the sibling group, not sponges.
Scientists discussed this finding for years, hampered by a lack of available methodologies to determine the right answer. “When DNA sequencing technologies developed, all the protocols were optimized for sequencing human genomes,” Schultz explains. “I had to figure out how to make this information work for ctenophores and sponges.”
In the last decade, these advancements gave scientists access to more comprehensive and precise information about genetic material. Schultz had access to the latest genomics research and discoveries because he was getting his PhD jointly at Monterey Bay Aquarium Research Institute and in the genomics department at the University of California, Santa Cruz. The latter has a rich legacy, as it was the department that first assembled the human genome. Schultz also studied with pioneering UC Santa Cruz biomolecular engineering professor Ed Green, a significant contributor to the Nobel Prize-winning work related to subsequent sequencing of the Neanderthal genome.
In his research, Schultz ultimately applied existing DNA sequencing technologies and new analysis techniques to organisms people hadn’t studied before. This enabled him to examine these genomes at the chromosome level. “In the past, when people said they sequenced the genome, they had little bits of pieces of DNA,” he says. “Think of it like if you took a book and tore all the pages out. You would have individual pages, but you wouldn’t know the order that they went in to make a chapter. But chromosome-scale is where you have the whole book, front to back, and you know the order that the pages go in.”
It’s like the longest game of telephone—or it’s the best handed-down
story of all time.”
This level of detail unlocked incredible insights. “Comparing the chromosome-scale genomes—the full stories—allowed us to figure out what events happened in the past,” Schultz says.
“We came up with methods to look at how pieces of DNA would fuse together and become mixed over time—and determined how that information could tell us how animals and other organisms are related to one another. People haven’t done that before for this specific type of genomic change that we looked at.”
Why are these findings so exciting? For starters, they change our understanding of the timing of animal evolution—more precisely, the evolution of animal-specific characteristics such as neurons, muscles, and genome organization. “This study tells us that the first neuron perhaps existed hundreds of millions of years earlier than it would have if sponges were the sister group of all animals,” Schultz says. “Maybe neurons were in things that were living in the ocean 700 million years ago.”
Schultz’s team also discovered that certain groups of linked genes were found on the same chromosomes in animals and close relations, like unicellular organisms. Incredibly, the genes’ location stayed the same—even as genes were passed down across the generations and as the chromosomal lineages of organisms and animals evolved independently from each other. “The common ancestor of the unicellular organisms and animals existed about a billion years ago,” Schultz explains, “which means that the genes we see linked together today existed on the same chromosome in that ancestor about a billion years ago.
“It’s like the longest game of telephone—or it’s the best handed-down story of all time.”
Schultz’s interest in studying DNA dates to his time at Oberlin. As a biology major, he cultivated his skills by pursuing research with two professors: Marta Laskowski, who studies plant root development, and Mike Moore, whose work involves plant evolutionary diversity.
“In Marta’s lab, everything that I did was centered around learning some technique with manipulating DNA,” Schultz says. “In Mike’s lab, I learned how DNA can tell us about how things are related.” This culminated in a senior thesis project with former professor Adam Haberman about the conservation of genes over hundreds of millions of years of evolution.
A decade later, Schultz calls his time at Oberlin critical to his development as a scientist—in no small part because the approach to research encouraged independence and a collaborative spirit.
“The professors are really passionate about making sure that students have chances to learn,” he says. “And they have the time to spend with the students. You have these brilliant people be your advisors in a way that you wouldn’t get in most other places around the world. Their guidance outside of the lab and their willingness to foster creativity, inquisitiveness, and the drive for knowledge was really inspiring. It made me really push myself to learn.”
Upon graduating, Schultz earned a Fulbright to study bioluminescent organisms and their DNA at Nagoya University in Japan. He continued his studies in Russia and earned a prestigious National Science Foundation Graduate Research Fellowship. As his career progressed, he discovered that it wasn’t just his Oberlin lab work that helped him become a better scientist.
A native of nearby Vermilion, Ohio, Schultz initially chose Oberlin to indulge his many interests outside of science—including art, music, and history. Accordingly, he pursued minors in chemistry and East Asian studies. “It ended up being the perfect place for me,” he says, “because I had a lot of freedom to explore all the intellectual pursuits that I wanted.”
Schultz’s Oberlin language studies paid off when he sought a Fulbright, which required him to complete his application in both English and Japanese. Over time, his broad skill set and varied interests also helped with research—like the ways he could apply the artistic skills he’s cultivated over the years to draw figures and take photos.
“Everything that you learn outside of your field comes together at some point,” he says. “And that’s how you can make big discoveries and do things that other people haven’t been able to do.”
Schultz hopes his research will lead to even greater things. The amount—and quality—of genomes published in the recent study are an invaluable resource for the evolutionary biology community.
“There aren’t many high-quality genomes of animals that are closely related to humans, worms, and arthropods—for example, of jellyfish, corals, sponges, ctenophores, and flat animals, or placozoans,” Schultz says. “The more genomes we sequence of things that are alive today, it gives us a clearer picture of what the ancestral genome was like a billion years ago.”
Sequencing more organisms is certainly on the plate. But Schultz’s research has also brought up additional intriguing questions to ponder. Why are the genes on animals still linked together after all this time—and how have they stayed connected for so long?
“There’s a question of whether they have stayed linked together for so long because they’re functionally linked—so if they were split up, the animal might die, or it might not pass its genes on,” he says. “Or maybe it’s due to random chance. That’s the next big question to ask of this finding.”



MBARI's remote operational vehicle Doc Ricketts is constantly on the search for comb Jellies across the California coast, where they like to dwell at depths of over 1,000 meters.
Photo credits: (top left) A California sea gooseberry (Hormiphora californensis) photographed in the laboratory; Darrin Schultz © 2021 MBARI. (bottom row, front to back) A lobed comb jelly (Bolinopsis microptera) observed by MBARI's remotely operated vehicle (ROV) Doc Ricketts offshore of Central California at a depth of approximately 1,640 meters; © 2021 MBARI. A lobed comb jelly (Bolinopsis microptera) observed by MBARI's remotely operated vehicle (ROV) Doc Ricketts offshore of Central California at a depth of approximately 1,360 meters; © 2017 MBARI. A lobed comb jelly (Bolinopsis microptera) observed by MBARI's remotely operated vehicle (ROV) Doc Ricketts at the Santa Lucia Knoll, offshore of Central California, at a depth of approximately 1,450 meters; © 2021 MBARI.
Annie Zaleski is the editor of the Oberlin Alumni Magazine
This story originally appeared in the Fall 2023 issue of the Oberlin Alumni Magazine.
Tags:
You may also like…
Voices Carry
March 13, 2025
Grammy-certified vocalist and viola da gamba player Ari Mason ’14 finds her niche in video games, films, and a vocal library.
London Calling
March 13, 2025
For Tracy Chevalier ’84, the Oberlin-In-London program was a magical, intense period of cultural and intellectual stimulation. As the beloved study-away experience celebrates 50 years, the New York Times best-selling author looks back on the semester she spent studying and living in London.
A Banner Held High
February 26, 2025
In 2018, Oberlin College named its main library after civil rights leader Mary Church Terrell, Class of 1884.


